Building a real-time big data pipeline (2: Spark Core, Hadoop, Scala)
Published:
Updated on May 07, 2020
Apache Spark is a general-purpose, in-memory cluster computing engine for large scale data processing. Spark can also work with Hadoop and its modules. The real-time data processing capability makes Spark a top choice for big data analytics.
The spark core has two parts. 1) Computing engine and 2) Spark Core APIs.
Spark provides APIs in Java, Scala, Python and R 1. It also supports libraries such as Spark SQL for structured data processing, MLlib for machine learning, GraphX for computing graphs, and Spark Streaming for stream computing.
Apache Spark Ecosystem
+--------+-----------+-------+----------+
| SQL | Streaming | MLlib | GraphX |
|---------------------------------------|
| Spark Core API |
|---------------------------------------|
| Scala | Python | Java | R |
|---------------------------------------|
| Compute Engine |
+---------------------------------------+
Spark Computing Engine: Hadoop MapReduce vs Spark
Apache Hadoop2 offers distributed storage (HDFS), resource manager (YARN) and computing framework (MapReduce). MapReduce reads and writes from disk, which slows down the processing speed and overall efficiency.
Apache Spark is a distributed processing engine comes with it’s own Spark Standalone cluster manager. However, we can also plugin a cluster manager of our choice such as YARN (the resource manager in Hadoop), Apache Mesos, or Kubernetes. When Spark applications run on YARN, resource management, scheduling, and security are controlled by YARN. Similarly, for the storage system we can use Hadoop’s HDFS, Amazon S3, Google cloud storage or Apache Cassandra. The Spark compute engine provides some basic functionality like memory management, task scheduling, fault recovery and most importantly interacting with the cluster manager and storage system. Spark also has a local mode, where the driver and executors run as threads on your computer instead on a cluster, which is useful for developing your applications from a personal computer. Spark runs applications up to 100 times faster in memory and 10 times faster on disk than Hadoop by reducing the number of read-write operations to disk and storing intermediate data in-memory3.
Spark Core APIs There are 3 alternatives to hold data in Spark. 1) Data Frame 2) Data Set and 3) RDD (Resilient Distributed Dataset). Data Frame and Data Set operate on structured data while RDD operate on unstructured data 4. The core APIs (available as Scala, Java, Python and R) facilitate the execution of high-level operators with RDD. RDD allows Spark to transparently store data on the memory, and send to disk only what’s important or needed. As a result, a lot of time that is spent on the disc read and write is saved.
Spark libraries
Spark libraries such as Spark SQL, Spark Streaming, MLlib and Graphx directly depend on Spark Core APIs to achieve distributed processing.
Figure. Spark is fully compatible with the Hadoop eco-system and works smoothly with HDFS (https://towardsdatascience.com)
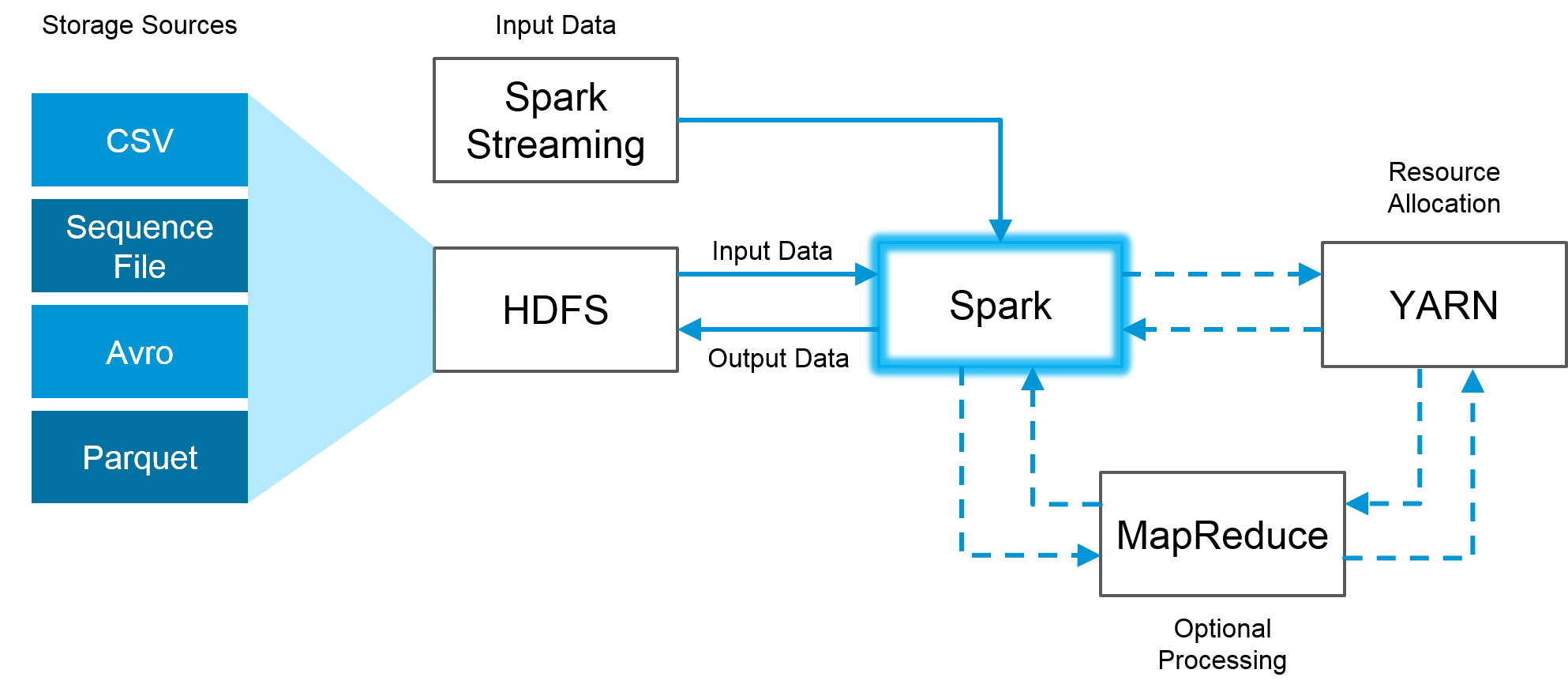
We can create RDDs using one of the two methods. 1) Load data from a source or 2) Create an RDD by transforming another RDD.
RDD: Resilient Distributed Dataset
Spark RDD is a resilient, partitioned, distributed and immutable collection of data4.
Resilient – RDDs are fault tolerant. If any bug or loss found, RDD has the capability to recover the loss.
Partitioned – Spark breaks the RDD into smaller chunks of data. These pieces are called partitions.
Distributed – Instead of keeping these partitions on a single machine, Spark spreads them across the cluster.
Immutable – Once defined, you can’t change them i.e Spark RDD is a read-only data structure.
For “RDDs vs DataFrames and Datasets - When to use them and why”, see reference 5.
Step 1: Hadoop installation
See tutorials on
Update your ~/.bash_profile file, which is a configuration file for configuring user environments.
$vi ~/.bash_profile
export HADOOP_HOME=/Users/adinasarapu/Documents/hadoop-3.1.3
export PATH=$PATH:$HADOOP_HOME/bin
Start Hadoop
start-dfs.sh - Starts the Hadoop DFS daemons, the namenode and datanodes. Use this before start-mapred.sh
stop-dfs.sh - Stops the Hadoop DFS daemons.
start-mapred.sh - Starts the Hadoop Map/Reduce daemons, the jobtracker and tasktrackers.
stop-mapred.sh - Stops the Hadoop Map/Reduce daemons.
$bash sbin/start-dfs.sh
Starting namenodes on [localhost]
Starting datanodes
Starting secondary namenodes [Ashoks-MacBook-Pro.local]
Verify Hadoop installation
$ jps
61073 ResourceManager
82025 SecondaryNameNode
61177 NodeManager
81882 DataNode
82303 Jps
81774 NameNode
Create user
$hadoop fs -mkdir -p /user/adinasarapu
Move file to HDFS (Hadoop Distributed File System)
$hadoop fs -copyFromLocal samples.csv /user/adinasarapu
Now the data file is at HDFS distributed storage. The file location at HDFS is hdfs://localhost:9000/user/adinasarapu/samples.csv
List files moved
$ hadoop fs -ls /user/adinasarapu
Found 3 items
-rw-r--r-- 1 adinasarapu supergroup 110252495 2020-02-08 17:04 /user/adinasarapu/flist.txt
-rw-r--r-- 1 adinasarapu supergroup 1318 2020-02-09 14:47 /user/adinasarapu/samples.csv
-rw-r--r-- 1 adinasarapu supergroup 303684 2020-02-09 08:21 /user/adinasarapu/survey.csv
Step 2: Apache Spark installation
For basic configuration see tutorial on Installing Apache Spark … on macOS
Update your ~/.bash_profile file, which is a configuration file for configuring user environments.
$vi ~/.bash_profile
export SPARK_HOME=/Users/adinasarapu/Documents/spark-3.0.0-preview2-bin-hadoop3.2
export PATH=$PATH:$SPARK_HOME/bin
Start Spark shell
$spark-shell
20/02/09 15:03:23 ..
Using Spark's default log4j profile: org/apache/spark/log4j-defaults.properties
Setting default log level to "WARN".
To adjust logging level use sc.setLogLevel(newLevel). For SparkR, use setLogLevel(newLevel).
Spark context Web UI available at http://192.168.0.5:4040
Spark context available as 'sc' (master = local[*], app id = local-1581278612106).
Spark session available as 'spark'.
Welcome to
____ __
/ __/__ ___ _____/ /__
_\ \/ _ \/ _ `/ __/ '_/
/___/ .__/\_,_/_/ /_/\_\ version 3.0.0-preview2
/_/
Using Scala version 2.12.10 (Java HotSpot(TM) 64-Bit Server VM, Java 1.8.0_102)
Type in expressions to have them evaluated.
Type :help for more information.
Read data from distributed storage (HDFS): csv file
scala> val df = spark.read.options(Map(
"header" -> "true",
"inferSchema"->"true",
"nullValue"->"NA",
"timestampFormat"->"MM-dd-yyyy",
"mode"->"failfast")).csv("hdfs://localhost:9000/user/adinasarapu/samples.csv")
Check the file for content
scala> df.show
+------+--------+---+-----+------+-------+-------+---------+-------+
|Sample| p16|Age| Race| Sex|Anatomy|Smoking|Radiation| Chemo|
+------+--------+---+-----+------+-------+-------+---------+-------+
|GHN-48|Negative| 68|white|female| BOT|current| Y| Y|
|GHN-57|Negative| 50|white|female| BOT|current| Y| Y|
|GHN-62|Negative| 71|white| male| BOT| never| Y| N|
|GHN-76|Negative| 60| AA| male| Tonsil| former| N| N|
|GHN-39|Positive| 51|white| male| BOT| never| Y| Y|
|GHN-60|Positive| 41|white| male| BOT| never| Y| Y|
|GHN-64|Positive| 49|white| male| BOT| never| Y| Y|
|GHN-65|Positive| 63|white| male| BOT| former| Y| Y|
|GHN-69|Positive| 56|white| male| BOT|current| Y| Y|
|GHN-70|Positive| 68|white| male| BOT| former| Y| Y|
|GHN-71|Positive| 59|white| male| BOT| never| N| Y|
|GHN-77|Positive| 53| AA| male| BOT| never| N| N|
|GHN-82|Positive| 67|white| male| BOT| former| Unknown|Unknown|
|GHN-43|Positive| 65|white| male| BOT| former| Y| Y|
|GHN-73|Positive| 72|white|female| Tonsil| never| Y| Y|
|GHN-40|Positive| 66|white| male| Tonsil| former| Y| Y|
|GHN-66|Positive| 52|white| male| Tonsil|current| Y| Y|
|GHN-67|Positive| 54|white| male| Tonsil| never| Y| Y|
|GHN-68|Positive| 51|white| male| Tonsil| former| Y| Y|
|GHN-79|Positive| 65|white| male| Tonsil| former| N| Y|
+------+--------+---+-----+------+-------+-------+---------+-------+
only showing top 20 rows
Check the number of partitions
scala> df.rdd.getNumPartitions
res4: Int = 1
Set/increase the number of partitions to 3
scala> val df2 = df.repartition(3).toDF
Recheck the number of partitions
scala> df2.rdd.getNumPartitions
res5: Int = 3
SQL like operation
Data Frame follows row and column structure like a database table. Data Frame compiles down to RDDs. RDDs are immutable; once loaded you can’t modify it. However, you can perform Transformations and and Actions. Spark Data Frames carries the same legacy from RDDs. Like RDDs, Spark Data Frames are immutable. You can perform transformation and actions on Data Frames.
scala> df.select("Sample","Age","Sex","Anatomy").filter("Age < 55").show
+------+---+------+-------+
|Sample|Age| Sex|Anatomy|
+------+---+------+-------+
|GHN-57| 50|female| BOT|
|GHN-39| 51| male| BOT|
|GHN-60| 41| male| BOT|
|GHN-64| 49| male| BOT|
|GHN-77| 53| male| BOT|
|GHN-66| 52| male| Tonsil|
|GHN-67| 54| male| Tonsil|
|GHN-68| 51| male| Tonsil|
|GHN-80| 54| male| Tonsil|
|GHN-83| 54| male| Tonsil|
+------+---+------+-------+
scala> df.groupBy('Sex).agg(Map("Age" -> "avg")).show
+------+------------------+
| Sex| avg(Age)|
+------+------------------+
|female|63.333333333333336|
| male| 58.125|
+------+------------------+
scala> val df1 = df.select("Sex","Radiation")
scala> df1.show
+------+---------+
| Sex|Radiation|
+------+---------+
|female| Y|
|female| Y|
| male| Y|
| male| N|
| male| Y|
| male| Y|
| male| Y|
| male| Y|
| male| Y|
| male| Y|
| male| N|
| male| N|
| male| Unknown|
| male| Y|
|female| Y|
| male| Y|
| male| Y|
| male| Y|
| male| Y|
| male| N|
+------+---------+
only showing top 20 rows
scala> val df2 = df1.select($"Sex",
(when($"Radiation" === "Y",1).otherwise(0)).alias("Yes"),
(when($"Radiation" === "N",1).otherwise(0)).alias("No"),
(when($"Radiation" === "Unknown",1).otherwise(0)).alias("Unknown"))
scala> df2.show
+------+-------+-------+-----------+
| Sex| Yes| No| Unknown|
+------+-------+-------+-----------+
|female| 1| 0| 0|
|female| 1| 0| 0|
| male| 1| 0| 0|
| male| 0| 1| 0|
| male| 1| 0| 0|
| male| 1| 0| 0|
| male| 1| 0| 0|
| male| 1| 0| 0|
| male| 1| 0| 0|
| male| 1| 0| 0|
| male| 0| 1| 0|
| male| 0| 1| 0|
| male| 0| 0| 1|
| male| 1| 0| 0|
|female| 1| 0| 0|
| male| 1| 0| 0|
| male| 1| 0| 0|
| male| 1| 0| 0|
| male| 1| 0| 0|
| male| 0| 1| 0|
+------+-------+-------+-----------+
only showing top 20 rows
scala>df2.groupBy("Sex").agg(Map("Yes" -> "sum", "No" -> "sum", "Unknown" -> "sum")).show
+------+--------+-------+------------+
| Sex|sum(Yes)|sum(No)|sum(Unknown)|
+------+--------+-------+------------+
|female| 3| 0| 0|
| male| 18| 5| 1|
+------+--------+-------+------------+
Scala user-defined function (UDF)
def parseSex(g: String) = {
g.toLowerCase match {
case "male" => "Male"
case "female" => "Female"
case _ => "Other"
}
}
scala> val parseSexUDF = udf(parseSex _)
scala> val df3 = df2.select((parseSexUDF($"Sex")).alias("Sex"),$"Yes",$"No",$"Unknown")
scala> val df4 = df3.groupBy("Sex").agg(sum($"Yes"), sum($"No"), sum($"Unknown"))
scala> df4.show
+------+------------+------------+----------------+
| Sex| sum(Yes)| sum(No)| sum(Unknown)|
+------+------------+------------+----------------+
|Female| 3| 0| 0|
| Male| 18| 5| 1|
+------+------------+------------+----------------+
scala> val df5 = df4.filter($"Sex" =!= "Unknown")
scala> df5.collect()
scala> df5.show
+----+------------+------------+----------------+
| Sex| sum(Yes)| sum(No)| sum(Unknown)|
+----+------------+------------+----------------+
|Male| 18| 5| 1|
+----+------------+------------+----------------+
Command to stop the interactive shell in Scala:
scala> Ctrl+D
References
RDDs vs Data Frames and Data Sets A Tale of Three Apache Spark APIs: RDDs vs DataFrames and Datasets - When to use them and why ↩
